


|

| ||
| teem | / | nrrd |
Example NRRD files |
This is the minimalist header for 2-dimensional data. Every NRRD file starts with the magic "NRRD0001" to unambiguously identify the file as a NRRD file, for NRRD readers, as well as any other readers that have to handle multiple formats. In this case, the dimension field says that this is two-dimensional data, and the sizes field is saying that the array has 128 samples along both its axes.NRRD0001 type: unsigned char dimension: 2 sizes: 128 128 encoding: raw WYXWYWVUVUSSTSPOPOOLKMJJJHFFFDCA data ... data ...
If we wanted to create a NRRD header which referred to the PGM image, but didn't contain any data itself, we could write this, which is the entire detached header:
Assuming that fool.pgm is in the same directory as this header file, the detached header is as good as fool.nrrd, usable from any other working directory.NRRD0001 type: unsigned char dimension: 2 sizes: 128 128 encoding: raw data file: ./fool.pgm line skip: 3
With either detached or attached headers, its easy to add comments to the header. They start with "#" and go to the line termination, and can be anywhere after the magic:
The content field is effectively a comment, but is intended to serve as a concise textual description of the contents of the NRRD file.NRRD0001 content: freshman year foolishness in Risley Hall type: unsigned char dimension: 2 # the original size was much larger sizes: 128 128 encoding: raw data file: ./fool.pgm # this skips past the PPM header, more lines would have to be # skipped if there where comments in the PPM file itself. line skip: 3
But so far this has just been an exercise to demonstrate the NRRD header. For data that can be stored in a PPM or PGM image, you might as well use PGM and PPM images, since the nrrd library can natively read and write PGM and PPM images with the exact same nrrdLoad() and nrrdSave() library calls as for NRRD files.
If the type of the data is 32-bit float instead of unsigned char, then byte ordering (endianness) information is required in the header: foolf.nrrd
If this had been written in a linux, cygwin, or windows box, the endian line would instead say "endian: little". NRRD readers know how to read either byte ordering, and no particular byte ordering is favored in the NRRD format. Other possible types include "int", "short", and "double".NRRD0001 type: float dimension: 2 sizes: 128 128 endian: big encoding: raw raw data ... data ...
In NRRD, whenever a field is giving information about each and every field (called "per-axis field"), the ordering of the axes is always fastest to slowest.NRRD0001 type: unsigned char dimension: 3 sizes: 3 128 128 encoding: raw X[JXYIWXFWXIXYIWVFV ... data ... data ...
Nearly all the field identifiers and descriptors are case-insensitive. The header above could just as well be:
NRRD0001 TYPE: UNSIGNED CHAR DIMENSION: 3 SIZES: 3 128 128 ENCODING: RAW X[JXYIWXFWXIXYIWVFV ... data ... data ...
Except for the fact that the per-axis fields must follow the dimension field, the fields can appear in any order. Also, some of the fields, (such as type) admit alternatives, so this is valid and equivalent:
NRRD0001 dimension: 3 encoding: raw sizes: 3 128 128 type: uchar X[JXYIWXFWXIXYIWVFV ... data ... data ...
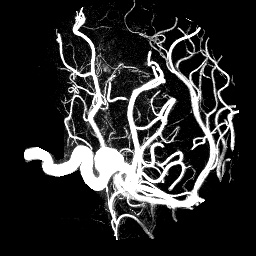
For instance, here's a header for aneurism.raw.gz. If your web browser decides to be helpful and uncompress the file as part of downloading, then use snarf to get the URL directly.
Except for the comment noting the original source of the data, a functionally identical header can be generated with one invocation of unu make -h:NRRD0001 content: aneurism # Courtesy of Philips Research, Hamburg, Germany dimension: 3 type: uchar sizes: 256 256 256 spacings: 1 1 1 data file: ./aneurism.raw.gz encoding: gzip
We record spacings for this dataset because unlike the previous examples, we know that information with certainty now. NRRD never forces you to represent information you don't actually know. The encoding field is what identifies this as gzip compressed data. This is fully inter-operable with the command-line gzip/gunzip tools. In this case, compression allows the dataset to be fifty times smaller, and the ability to leave data compressed is very conveniant.unu make -h -i ./aneurism.raw.gz -t uchar -s 256 256 256 -sp 1 1 1 \ -c aneurism -e gzip -o aneur.nhdr
If we save the header above to a file aneurism.nhdr, in the same directory as the aneurism.raw.gz file, then we're done. We can immediately start using unu, for instance, to start inspecting the data:
unu project -i aneurism.nhdr -a 0 -m max -o an-max.pgm
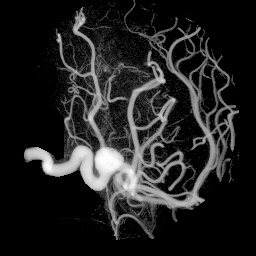
unu project -i aneurism.nhdr -a 0 -m variance | unu gamma -g 4 \ | unu quantize -b 8 -o an-var.pgm
The axis mins and axis maxs fields are used to represent the range of values which were mapped to the lowest and highest bins of the histogram. Because the bins of the histogram are necessarily cell-centered, this is recorded via the centers field. unu also tries to generate a consise verbal description of the data it produces, based on the content field of the input. In this case, the new content was also used in the labels field, which allows arbitrary strings to be associated with each axis.unu histo -i aneurism.nhdr -b 256 | unu save -f nrrd -e ascii -o an-hist.nrrd NRRD0001 content: histo(aneurism,256) type: unsigned int dimension: 1 sizes: 256 axis mins: 0 axis maxs: 255 centers: cell labels: "histo(aneurism,256)" encoding: ascii 16608268 3600 3506 3380 3097 3197 ...